Save the Date - New CAMPI Webinar
Monday, Dec 15, 2025 9–11 a.m. EST / 3–5 p.m. CET (7:30–9:30 p.m. Mumbai / 10 p.m.–12 a.m. Shangai)
Register here for the webinar.
If you’d like to be part of an upcoming CAMPI, you can already express your interest here!

7th International Metaproteomics Symposium

Visit the website for more details!
We encourage all members to promote the event by adding the following official email signature to their professional communications (HTML code available here).
Executive Board Digest
Call for Submissions: Metaproteomics Initiative’s Best Manuscript Award 2026
The Metaproteomics Initiative is excited to announce the Best Manuscript Award 2026! This award recognizes early career researchers (ECRs) for outstanding and innovative contributions to metaproteomics.
Submission Deadline: March 1, 2026 Award Announcement: IMS 2026 Eligibility: ECRs (PhD not yet obtained or awarded after June 1, 2020) who have submitted an original research manuscript to a peer-reviewed journal or preprint server since November 1, 2024.
Don’t miss the opportunity to showcase your research and gain international recognition within the metaproteomics community! Check this website for more information on how to submit.
Your Community
METAMIC³ - Advancing Microbiome Science Through Metaproteomics
The METAMIC³ project unites leading academic and industry partners to train a new generation of researchers in metaproteomics and microbiome science. The program tackles One Health challenges through an integrated approach, from microbial mechanisms and ecosystem dynamics to clinical and biotechnological applications.
Some Doctoral Candidate (DC) positions are still open! (DC15) PhD student (m/f/d): Metabolic Labeling of Complex Microbial Communities
Community Feedback on New SDRF-Proteomics Terms for Metaproteomics
As presented at the IMS meeting in Oslo, the current SDRF-Proteomics terms don’t fully capture microbial environments. During his stay at the European Bioinformatics Institute (EMBL-EBI), Tim Van Den Bossche has been developing an extended set of metadata terms to improve metaproteomics data description.
We now invite the metaproteomics community to share feedback on the proposed terms and help identify which should be required for different environments. Your input will help shape the next version of SDRF-Proteomics and ensure it meets the needs of the metaproteomics community!
Contribute your feedback Read the draft guidelines
Metaproteomics articles you don’t want to miss
The Microbiologist’s Guide to Metaproteomics
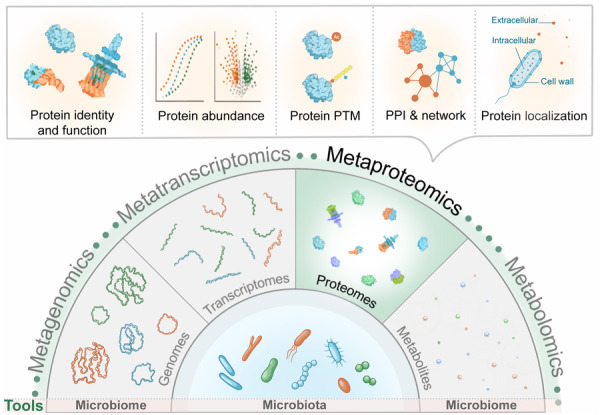
Developed by the Metaproteomics Initiative, this review is a practical guide to using metaproteomics to explore microbiome function. It explains how protein-level profiling reveals active processes and ecological roles within microbial communities, and how integrating metaproteomics with other omics provides a comprehensive view of microbial dynamics.
The review summarizes key principles, state-of-the-art methods, and analytical workflows, from experimental design and sample prep to mass spectrometry, data analysis, and statistics, serving both microbiome and proteomics researchers.
Benchmarking Fecal Sample Stabilization for Metaproteomics: Insights from CAMPI-2
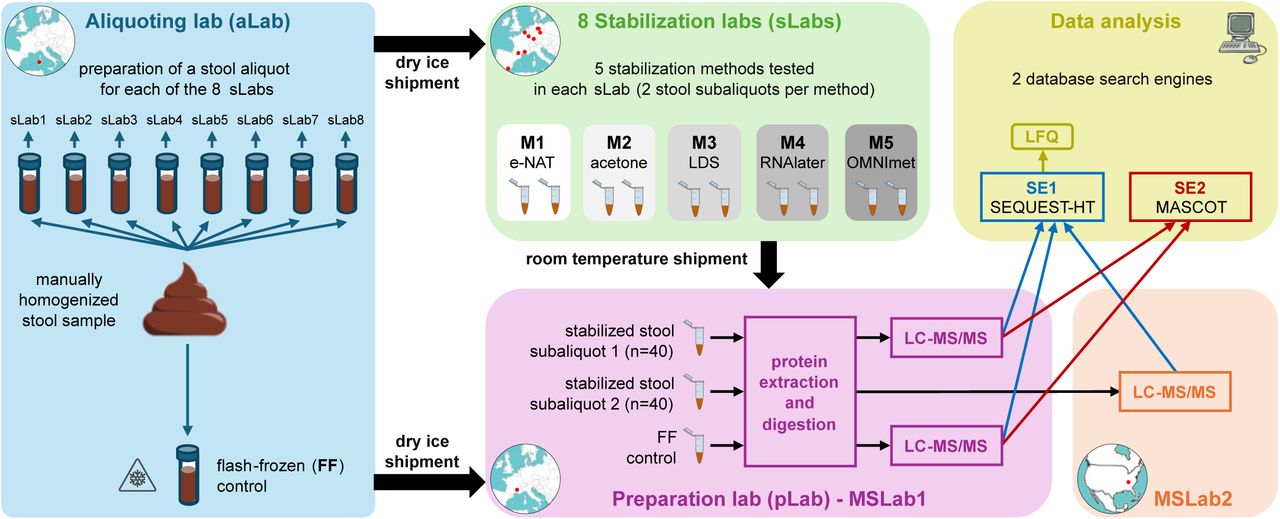
Reliable fecal metaproteomics requires preserving samples from collection to analysis—especially when immediate freezing isn’t possible. The CAMPI-2 multicenter study tested five stabilization approaches (two commercial kits; three reagent protocols) across eight European labs during room-temperature storage and shipment.
Stabilization choice substantially influenced taxonomic/functional profiles and reproducibility. A swab-based commercial device delivered the most consistent performance overall. Findings provide practical guidance for metaproteomic and multi-omic studies conducted under non-ideal storage conditions.
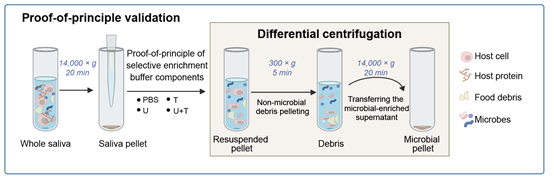
OSaMPle workflow for salivary metaproteomics analysis reveals dysbiosis in inflammatory bowel disease patients
The human oral microbiome is increasingly linked to inflammatory conditions such as IBD, yet probing its functional dynamics remains challenging due to high host protein content and low microbial biomass in oral samples. OSaMPle, an Optimized Salivary MetaProteomic workflow, enriches bacterial cells and reduces host interference in saliva and mouth-rinse.
Compared to a conventional approach, OSaMPle increased identification of bacterial peptides 3.2-fold and proteins 1.7-fold. Applied to IBD mouth-rinse samples, it revealed disease-specific signatures: reduced Peptostreptococcus fatty-acid elongation proteins and elevated Neisseria TCA-cycle proteins. The workflow is compatible with small volumes and high-throughput automation, offering a powerful tool for functional profiling of the oral microbiome.
Metaproteomics in the One Health Framework for Unraveling Microbial Effectors in Microbiomes
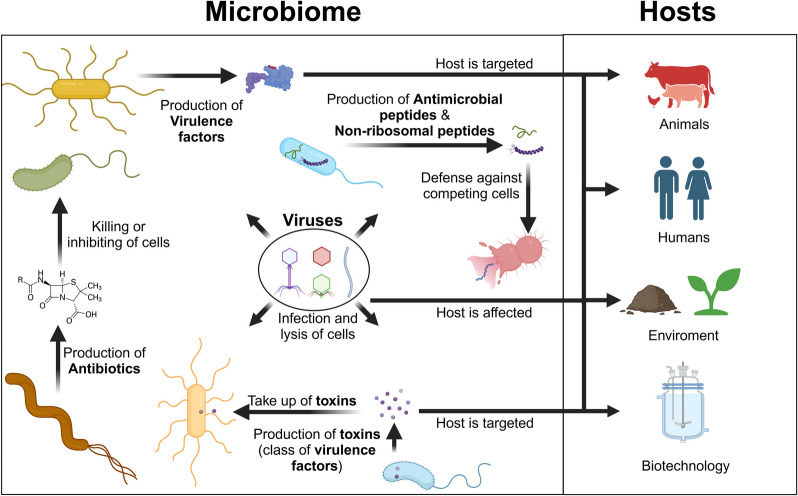
One Health seeks to integrate and balance the health of humans, animals, and environmental systems, which are intricately linked through microbiomes. These microbial communities exchange microbes and genes, influencing human and animal health as well as environmental, agricultural, and biotechnological processes.
Preventing pathogen emergence, and monitoring or controlling microbiome composition via microbial effectors (e.g., virulence factors, toxins, antibiotics, non-ribosomal peptides, viruses), holds transformative potential. Yet, how these effectors shape microbiomes and impact host and ecosystem health remains poorly understood.
Metaproteomics offers a powerful framework by quantifying microbial biomass composition and metabolic functions, and by detecting effectors such as viruses, antimicrobial resistance proteins, and non-ribosomal peptides. Here, we highlight how metaproteomics can elucidate microbial effectors and their impacts, and discuss opportunities to modulate microbiomes toward desired functions.
Ultra-sensitive metaproteomics redefines the dark metaproteome, uncovering host–microbiome interactions and drug targets in intestinal diseases

The functional analysis of host–gut microbiome interactions has been limited by the sensitivity of existing metaproteomic methods. uMetaP, an ultra-sensitive workflow combining advanced LC–MS technology with de novo sequencing (novoMP), expands functional coverage and increases detection of low-abundance microbial and host proteins by up to 5000-fold. Applied to models of intestinal injury and Crohn’s disease, uMetaP uncovered key host–microbiome functional networks and introduced the concept of a druggable metaproteome, providing new opportunities for therapeutic discovery.
Consumption of traditional Sardinian fermented milk promotes changes in the rat gut microbiota composition and functions
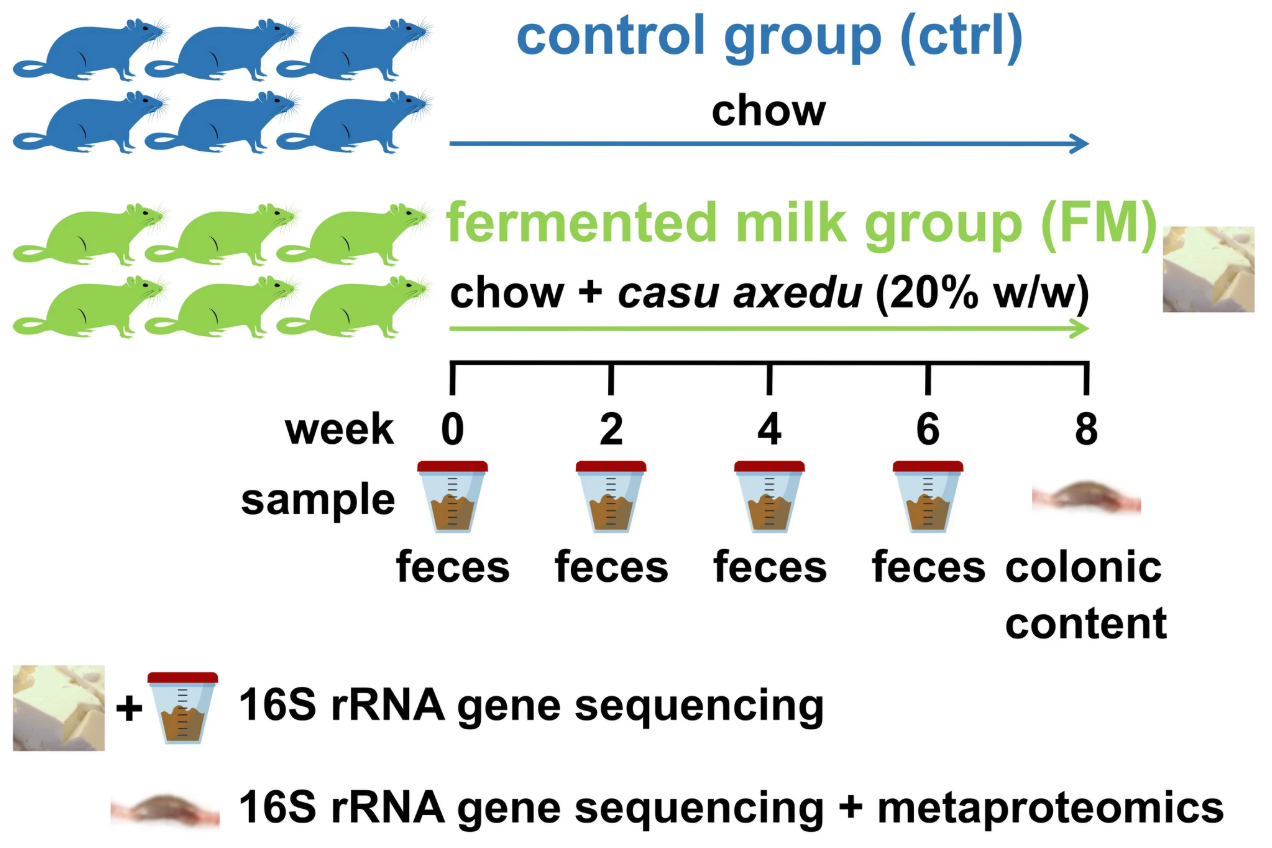
Fermented milk is a staple of Mediterranean diets, often enriched with lactic acid bacteria and fermentation-derived metabolites. Using an integrated 16S rRNA gene-based taxonomic profiling and metaproteomics-based functional analysis, Abbondio et al., characterized the effects of casu axedu consumption on the gut microbiota of rats in a controlled experimental setting, assessing both taxonomic composition and functional activity.
Specifically, metaproteomics allowed the characterization of the molecular processes occurring in the colon microenvironment by the simultaneous quantification of gut microbiota and host proteins contained in the colonic luminal contents. Together, the findings suggest that casu axedu consumption promotes a healthier gut microbiota and reduced inflammation through functional microbial shifts.
Contribution of metaproteomics to unveiling the functional role of the gut microbiome in human physiology and metabolism
Current fecal metaproteomics provides an initial understanding of the roles of gut microbes in human health, revealing redundant and taxon-specific functions. Future research should prioritize standardization, large-scale studies, and integration with multi-omics to better understand HGM metabolism. Emerging technologies, advanced mass spectrometry platforms, and AI-driven analytics are expected to increase sensitivity and depth of gut metaproteomics, accelerating discovery and potential clinical applications.