Advancing Metaproteomics: Insights from the 6th International Symposium

The 6th International Metaproteomics Symposium took place from January 13–15, 2025, at the scenic Soria Moria Hotel in Holmenkollen, Oslo, Norway. Set against a breathtaking winter landscape, the venue provided a spectacular backdrop for scientific exchange and collaboration. This year’s symposium brought together 79 participants from 18 countries, fostering a vibrant exchange of ideas and expertise.
Discover the key highlights of the Symposium here.
Award Winners’ Take on the 6th Metaproteomics Symposium
Best talk award, Lior Lobel
As a first-time attendant of the IMS I felt star-struck by the incredible researchers, whose names I recognized from the numerous papers I read since jumping onto the field two years ago. Read Lior’s full take on the symposium.
Best poster award, Valkhari Kale
As an early career researcher, it was reassuring to find a community that truly understands the challenges and methodological gaps in metaproteomics. Discover Vaikhari’s insights here.
Travel grant, Patrick Hellwig
It was really interesting to see all the new trends in metaproteomics workflows, like de novo, isotopes, and DIA. It was also great to get a good overview of how useful metaproteomics is. Find out Patrick’s key takeaways.
Best poster award, Rajesh Bajiya
Attending the IMS in Oslo was a fantastic experience! As a doctoral student, I had the opportunity to engage in insightful discussions, connect with experts in the field, and follow current research trend. Learn more from Rajesh’s experience.
Travel grant, Benoit Kunath
The IMS has quickly become one of the events I’m looking forward the most. Learn more from Benoit’s experience
ISME Trailblazer Award, Grace DAngelo
I was very excited to attend the IMS6 this year in Oslo and grateful for the opportunity to present my work. At the beginning of my PhD, I had a strong interest in learning metaproteomics and the personal goal of attending at least one IMS meeting. Learn more from Grace’s experience
Best Manuscript Award

Alfredo and his co-workers applied an integrated metagenomics-metaproteomics approach to show that different dietary protein sources altered the composition and function of the gut microbiome of mice fed defined-diets with different dietary protein sources.
Discover more about this study and Alfredo’s research on our website.
Executive Board Digest
3 New CAMPIs (Critical Assessment of MetaProteome Investigation) are being launched and require your participation! More details coming soon on our website.
Interested in joining the next CAMPI? Let us know!
CAMPI-3: Taxonomic and Functional Annotation of Metaproteomics Data
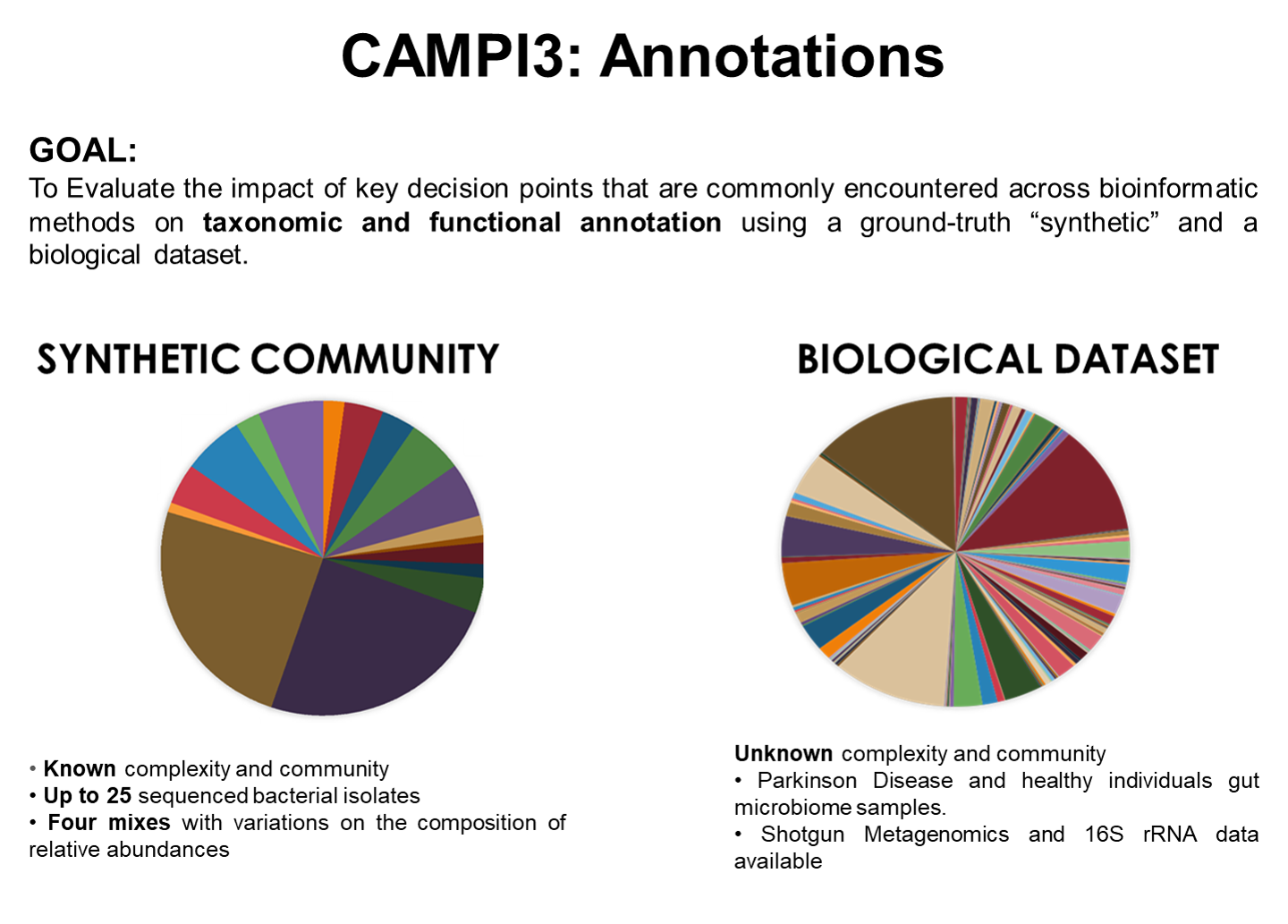
If you participated in CAMPI3, last November, you received the taxonomic lineage of each organism in the synthetic database. If you haven’t yet, please re-run your analysis (or complete your post-processing) using this additional information and share your updated results with the CAMPI3 team.
Having trouble finding the email from the CAMPI3 team? Contact them!
Help Shape the Metaproteomics Research Groups Page!
At the International Metaproteomics Symposium in Oslo, we received a request to expand our Research Groups page in two key ways:
- Adding a list of core facilities
- Labeling research groups by expertise
Before we move forward, we’d love your input to ensure this resource is truly valuable to the community!
Core Facilities
Would this be a useful resource for you? What details should be included beyond just listing the facilities? (e.g., available instruments, access options like collaboration or fee-for-service, other relevant info?)
Research Groups Page
Do you currently use it? Would adding labels or tags (e.g., bioinformatics, wet-lab, clinical applications, human gut, soil, etc.) make it more useful? If we add labels, would you actively use the page?
Since maintaining these resources takes effort, your honest feedback is essential! Share your thoughts with us by April 8, 2025.
Metaproteomics at the APT-2025, BRIC-ILS and BRIC-NCCS in India

Leyuan Li (Beijing Proteome Research Center, China), Magnus Artnzen (Norges miljø- og biovitenskapelige universitet, Norway) and Pratik Jagtap (University of Minnesota, USA) conducted a day-long workshop at the Advances in Proteomics Technologies (APT-2025) held at IIT Bombay (India). Read the recap here.
Save the Date
Applied Metaproteomics Workshop
The de.NBI course on Applied Metaproteomics is designed to train participants in the complete metaproteomics workflow, from experimental design and sample preparation to measurement and bioinformatics analysis of high-resolution mass spectrometry (MS) data. The course accommodates a maximum of 10 participants, who are encouraged to bring their own samples to begin their metaproteomics projects.
The workshop will be held at the Otto-von-Guericke University Magdeburg by Prof. Dirk Benndorf and Prof. Robert Heyer.
Find out how to register here.
EuPA 2025
Submitting an abstract for the EuPA 2025 conference in Saint-Malo? Does your abstract involve metaproteomics, and preferably a collaboration between several labs registered at the Metaproteomics Initiative? Next to the regular sessions, also tick the box for the EuPA initiatives full day session to be additionally considered for a talk at our own Metaproteomics Initiative session on Tuesday morning. Abstract submissions close on March 28, don’t miss the deadline!

Metaproteomics articles you don’t want to miss
Introducing PhyloFunc: A Phylogeny-Informed Functional Beta-Diversity Metric for Metaproteomics
Metaproteomics deserves a dedicated β-diversity metric In microbiome research, comparing functional profiles between microbial communities is essential for understanding ecosystem dynamics. Typically, β-diversity serves as one of the initial analytical steps of microbiome data analysis. However, when applying β-diversity analysis to metaproteomics, existing methods such as Bray-Curtis dissimilarity fail to capture the unique characteristics of protein-level functional relationships.
Why Do Conventional Methods Fall Short? Traditional beta-diversity metrics treat proteins as independent variables, ignoring the evolutionary and functional interconnections among them. This oversight disregards functional redundancy – different proteins performing similar roles in the community, and phylogenetic relatedness – proteins originating from closely related taxa often exhibit compensatory functional shifts.
The solution: PhyloFunc To bridge this gap, Wang and colleagues developed PhyloFunc, a phylogeny-informed functional distance metric that integrates, metaproteomic functional profiles, taxonomic origination of proteins and evolutionary relationships among taxa. Built upon the weighted UniFrac distance framework, PhyloFunc weighs functional differences by branch length in a phylogenetic tree, making it a biologically meaningful metric for metaproteomic beta-diversity. Key advantages of PhyloFunc include its ability to capture functional compensation by detecting shifts in function among phylogenetically related species, enhanced sensitivity in differentiating microbiome functional responses compared to traditional metrics, and its ready-to-use Python package, available on PyPI for seamless integration into your workflows. Get Started with PhyloFunc Today!
Insights on Wet and Dry Workflows for Human Gut Metaproteomics
The human gut microbiota (GM) is a community of microorganisms that resides in the gastrointestinal (GI) tract. Recognized as a critical element of human health, the functions of the GM extend beyond GI well-being to influence overall systemic health and susceptibility to disease. Among the other omic sciences, metaproteomics highlights additional facets that make it a highly valuable discipline in the study of GM. Indeed, it allows the protein inventory of complex microbial communities. Proteins with associated taxonomic membership and function are identified and quantified from their constituent peptides by liquid chromatography coupled to mass spectrometry analyses and by querying specific databases (DBs). The aim of this review was to compile comprehensive information on metaproteomic studies of the human GM, with a focus on the bacterial component, to assist newcomers in understanding the methods and types of research conducted in this field. The review outlines key steps in a metaproteomic-based study, such as protein extraction, DB selection, and bioinformatic workflow. The importance of standardization is emphasized. In addition, a list of previously published studies is provided as hints for researchers interested in investigating the role of GM in health and disease states.
Your Community
Center of Excellence for Metaproteomics
 The new Center of Excellence for Metaproteomics at the University of Vienna will focus on microbiome research using high-resolution protein-based methods. The strategic partnership between the University of Vienna and Bruker combines cutting-edge research with state-of-the-art technology. The new Center of Excellence for Metaproteomics will be headed by biochemist David Gómez-Varela from the University of Vienna. Read the press release.
The new Center of Excellence for Metaproteomics at the University of Vienna will focus on microbiome research using high-resolution protein-based methods. The strategic partnership between the University of Vienna and Bruker combines cutting-edge research with state-of-the-art technology. The new Center of Excellence for Metaproteomics will be headed by biochemist David Gómez-Varela from the University of Vienna. Read the press release.
Job Alert: HoLMiR seeks a research engineer
The Hohenheim Center for Livestock Microbiome Research seeks a research engineer.
The University of Hohenheim is one of the leading universities in agricultural science and the pioneer in microbiome research of Livestock in Germany. The new HoLMiR infrastructure will offer state-of-the-art research facilities. A central part is the mass spectrometry lab and offers qualitative and quantitative analyses of proteins and metabolites. We are looking for a technical manager to join our new team. Preferred starting date is June 1st, 2025 or later. Check all the details here.
Unipept 6.2.0: New Features for Metaproteomics Analysis
Version 6.2.0 of the Unipept Web application was released on March 24th and brings several exciting new features and enhancements, expanding its capabilities to match those of the Unipept Desktop application. With this latest version, users can now create custom databases on the web and perform metaproteomics analysis on subsets of the UniProtKB database. This includes the ability to filter by NCBI taxon IDs, UniProt reference proteomes, or UniProtKB protein accession numbers directly from the user-friendly interface.