From the co-administrators
Dear members of the Metaproteomics Initiative,
We’re once again delighted to bring you exciting news about the metaproteomic field. In particular, we are glad to announce that the registrations for the 6th IMS in Oslo are now open! On top of it, our new early career representative has come up with a new twist for the symposium with the first edition of the MPI best manuscript award! Check it out below for a chance at the spotlight and support to attend the IMS.
Finally, we received a great amount of publication’s digest for this newsletter. We’ve just had a look at it, it’s definitely a great time to be doing metaproteomics!
After the worldwide success of the webinar in June, we can only be excited to see many of you soon again, to engage, and to interact regarding Initiative and all its projects.
Kind regards, Tim Van Den Bossche and Benoit Kunath
Executive Board Digest
Metaproteomics Webinar 2024
The 2nd Metaproteomics Webinar, held on June 11, 2024, was an online extravaganza with a truly global feel.
We were thrilled to receive a range of fascinating abstracts fitting our themes of health and the environment. After a tough selection process, we showcased presentations from brilliant researchers all over the world.
The turnout was fantastic, with attendance peaking at over 100. Attendees got the chance to virtually “meet” the Metaproteomics Initiative board and catch exciting updates on the latest CAMPI (Critical Assessment of Metaproteome Investigation) projects.
The event was such a hit that we’re already planning more webinars to spread the word on metaproteomics even further.

Save the Date

First MPI Best Manuscript Award
We are excited to announce the first Metaproteomics Initiative Best Manuscript Award, designed to recognize the best manuscript in the field of metaproteomics.
Eligibility: All ECRs (those who have not yet been awarded a PhD or were awarded a PhD after January 1, 2019) are eligible to apply. Manuscripts must have been submitted to a journal or preprint server between the 5th IMS (April 2023 in Avignon) and November 1, 2024.
How to Apply: To participate, submit your manuscript along with a 300-word statement outlining the significance of your research to info@metaproteomics.org by November 1, 2024.
Selection Process: All manuscripts will be reviewed by our MPI Best Manuscript Award committee. The top three manuscripts will be shared with the metaproteomics community in early December. MPI members will then have the opportunity to vote for their favorite manuscript ahead of the 6th IMS in Oslo.
Award: The winner will have the unique opportunity to present their work in a dedicated session during the symposium and some financial support to attend it.
For more details and publication criteria, please visit our homepage If you have any questions, feel free to reach out to us at info@metaproteomics.org.
Metaproteomics articles you don’t want to miss
Ultra-sensitive metaproteomics redefines the dark field of metaproteome, enables single-bacterium resolution, and discovers hidden functions in the gut microbiome
Metaproteomics uniquely characterizes host-microbiome interactions. However, most species detected by metagenomics remain hidden to metaproteomics due to sensitivity limits.
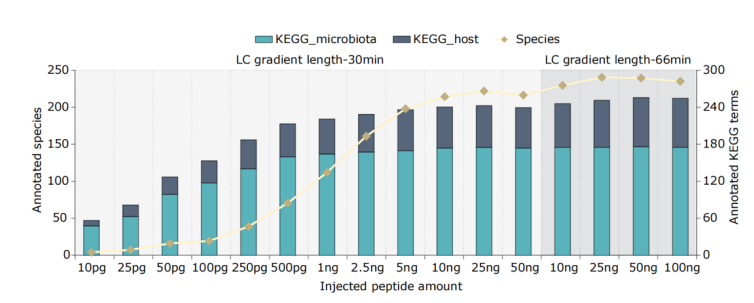
Feng Xian et al., from the group led by David Gomez-Varela, present a novel ultra-sensitive metaproteomic solution (uMetaP) that, for the first time, reaches full-length 16S rRNA taxonomic depth and can simultaneously decipher functional features. Querying the mouse gut microbiome, uMetaP achieved unprecedented performance in key metrics like protein groups (47925) alongside taxonomic (220 species) and functional annotations (223 KEGG pathways)-all within 30-min analysis time and with high reproducibility, sensitivity, and quantitative precision. uMetaP revealed previously unidentified proteins of unknown functions, small proteins, and potentially new natural antibiotics. Leveraging the extreme sensitivity of uMetaP and SILAC-labelled bacteria, they revealed the true limit of detection and quantification for the “dark” metaproteome of the mouse gut. Moreover, using a two-bacteria proteome mix, they demonstrated single-bacterium resolution (500 fg) with exceptional quantification precision and accuracy. From deciphering the interplay of billions of microorganisms with the host to exploring microbial heterogeneity, uMetaP represents a quantum leap in metaproteomics. Taken together, uMetaP will open new avenues for our understanding of the microbial world and its connection to health and disease.
Metaproteomic portrait of the healthy human gut microbiota
Gut metaproteomics can provide direct evidence of microbial functions actively expressed in the colonic environments, contributing to clarify the role of the gut microbiota in human physiology. In this study, Tanca et al., re-analyzed 10 fecal metaproteomics datasets of healthy individuals from different continents and countries, with the aim of identifying stable and variable gut microbial functions and defining the contribution of specific bacterial taxa to the main metabolic pathways.
The “core” metaproteome included 182 microbial functions and 83 pathways that were identified in all individuals analyzed. Several enzymes involved in glucose and pyruvate metabolism, along with glutamate dehydrogenase, acetate kinase, elongation factors G and Tu and DnaK, were the proteins with the lowest abundance variability in the cohorts under study. On the contrary, proteins involved in chemotaxis, response to stress and cell adhesion were among the most variable functions. Random-effect meta-analysis of correlation trends between taxa, functions and pathways revealed key ecological and molecular associations within the gut microbiota. The contribution of specific bacterial taxa to the main biological processes was also investigated, finding that Faecalibacterium is the most stable genus and the top contributor to anti-inflammatory butyrate production in the healthy gut microbiota. Active production of other mucosal immunomodulators facilitating host tolerance was observed, including Roseburia flagellin and lipopolysaccharide biosynthetic enzymes expressed by members of Bacteroidota. This study provides a detailed picture of the healthy human gut microbiota, contributing to unveil its functional mechanisms and its relationship with nutrition, immunity, and environmental stressors.
PathwayPilot: A User-Friendly Tool for Visualizing and Navigating Metabolic Pathways
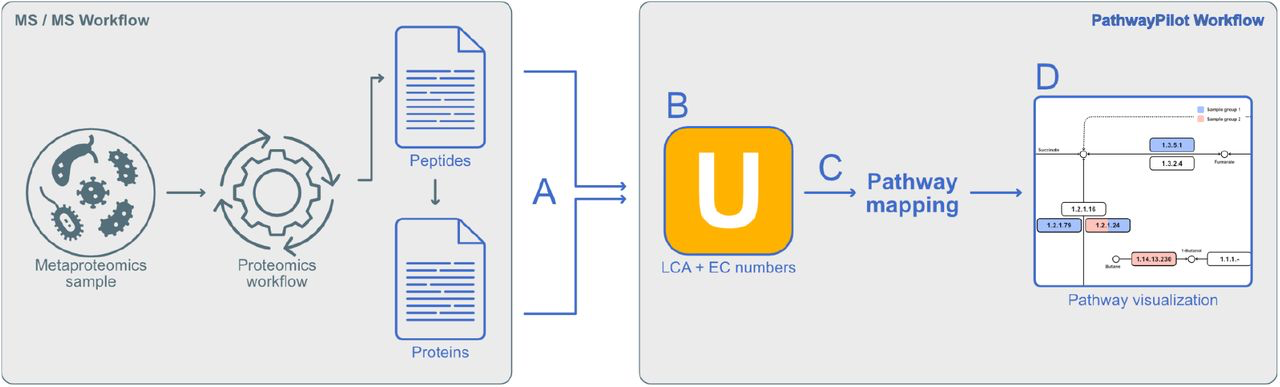
PathwayPilot is a new web application that simplifies the peptide- and protein-centric analysis and visualization of metabolic pathways in metaproteomics. It enables researchers to compare functions across different samples, offering insights into complex microbial communities.
A case study on the impact of caloric restriction on gut microbiota demonstrated the tool’s efficacy in deciphering complex metaproteomic data. The re-analysis revealed significant shifts in enzyme expressions related to short-chain fatty acid biosynthesis, aligning with existing research findings and showcasing PathwayPilot’s capability for accurate functional annotation and comparison across different microbial communities.
A novel clinical metaproteomics workflow enables bioinformatic analysis of host-microbe dynamics in disease
Clinical metaproteomics holds immense potential for expanding our understanding of host-microbe interactions underlying human disease. However, challenges persist in this field, with the foremost one being the characterization of microbial proteins present in low abundances relative to host proteins. Other hurdles faced by researchers stem from the use of very large protein sequences databases in peptide and protein identification from mass spectrometry (MS) data in addition to performing rigorous analyses of identified peptides, including taxonomic and functional assignments as well as statistical analysis to attain differentially abundant peptides.
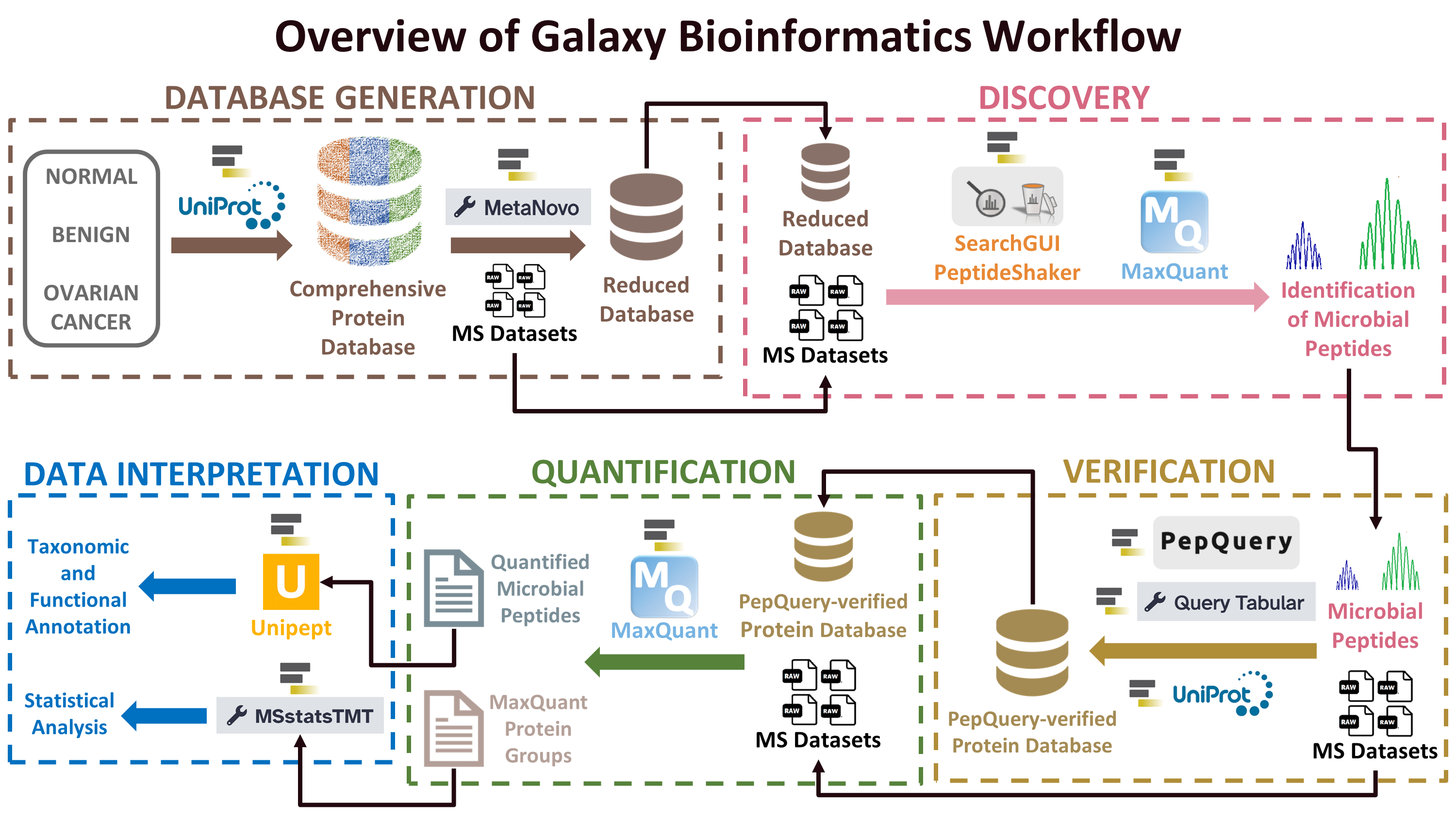
In a resource report recently published in mSphere, Katherine Do along with the Galaxy-P team are excited to present a clinical metaproteomics workflow that comprises custom database generation, peptide-spectrum match generation and verification, quantification, followed by taxonomic and functional assignments and statistical analysis. Not only does this workflow provide insight into host-microbe dynamics in disease via characterization of host proteins and prioritization of microbial proteins, it also leverages the computational power of the Galaxy bioinformatics platform during workflow development and execution, enabling us to perform compute-intensive analyses using large volumes of data. This clinical metaproteomics workflow has been used to characterize various sample types such as a) Metaproteomic Analysis of Nasopharyngeal Swab Samples to Identify Microbial Peptides in COVID-19 Patients (Bihani et al 2023) and b) for studying host-microbe dynamics in bronchoalveolar lavage samples applied to cystic fibrosis disease (Kruk et al 2024) and c) to generate a novel host and microbial biomarker panel for early detection of ovarian cancer (Mehta et al; ongoing work). We hope this workflow will prove useful as researchers strive to decipher pathogenic mechanisms of diseases, including cancer.
This workflow will also be part of the Proteomics section of the Galaxy Training Academy which is being conducted from October 7th to 11th. The Galaxy Academy is an online training event for Beginners as well as learners who would like to improve their Galaxy data analysis skills. Please register here if you are interested!
Benchmarking low- and high-throughput protein cleanup and digestion methods for human fecal metaproteomics
Fecal metaproteomics is an experimental approach that allows the investigation of gut microbial functions, which are involved in many different physiological and pathological processes. Standardization and automation of sample preparation protocols in fecal metaproteomics are essential for its application in large-scale studies.
Here, Tanca et al., comparatively evaluated different methods, available also in a high-throughput format, enabling two key steps of the metaproteomics analytical workflow (namely, protein cleanup and digestion). The results of this study provide critical information that may be useful for the optimization of metaproteomics experimental pipelines and their implementation in laboratory automation systems.
Biodiversity Analysis of Metaproteomics Samples with Unipept: A Comprehensive Tutorial
Metaproteomics has become a crucial omics technology for studying microbiomes. In this area, the Unipept ecosystem has emerged as a valuable resource for analyzing metaproteomic data. It offers in-depth insights into both taxonomic distributions and functional characteristics of complex ecosystems.
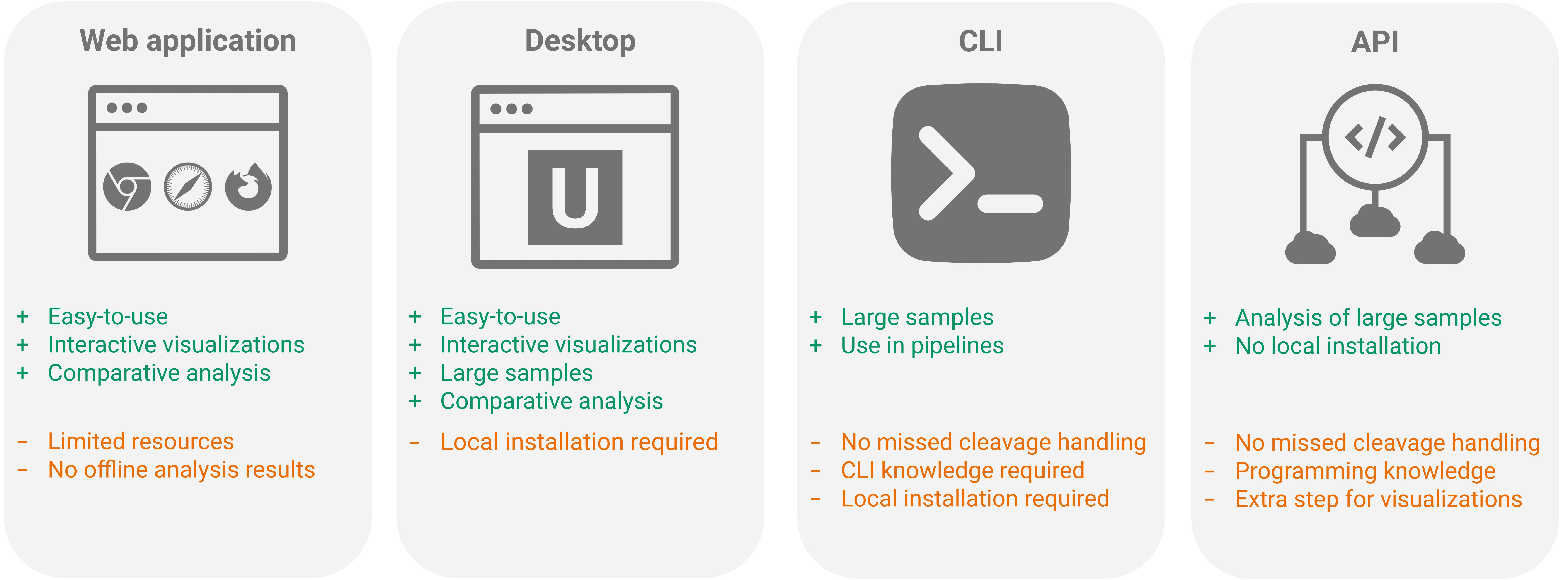
This tutorial explains essential concepts like Lowest Common Ancestor (LCA) determination and the handling of peptides with missed cleavages. It also provides a detailed, step-by-step guide on using the Unipept Web application and Unipept Desktop for thorough metaproteomics analyses. By integrating theoretical principles with practical methodologies, this tutorial empowers researchers with the essential knowledge and tools needed to fully utilize metaproteomics in their microbiome studies.